Laboratory
Reverse Transcriptase – qPCR Test: SARS-CoV-2 in Wastewater Samples
Apr. 1 2021
Bureau Veritas offers a test that quantifies the SARS-CoV-2 virus in wastewater samples. The method uses a 1-step reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) method.
The SARS-CoV-2 virus partitions predominantly to solids, rather than the aqueous phase. By targeting the solids in a wastewater sample, the test provides a more sensitive and representative result.
Sample Collection and Preservation
Samples are collected in 250 mL containers with no preservative. Collected samples should be immediately refrigerated and laboratory processing should begin within 48 hours of sample collection.
Genetic material from the SARS-CoV-2 virus is prone to degradation in wastewater within the first 48 hours after collection, if stored at room temperature. Refrigeration of wastewater samples can minimize the loss of genetic material for up to 4 to 5 days.
SAMPLE PREPARATION AND EXTRACTION
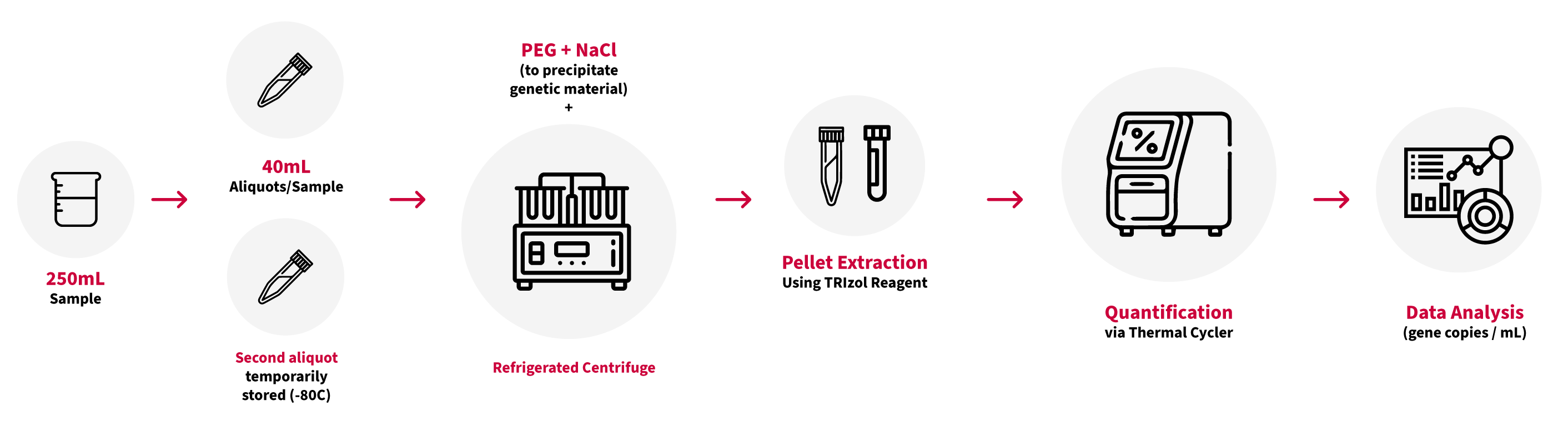
The contents of each 250 mL container are homogenized. Two 40mL aliquots are poured into separate tubes.
One aliquot is processed for analysis, while the second is stored in a freezer, should the sample require retesting. The 40mL aliquot submitted to the laboratory is processed using polyethylene glycol (PEG) and sodium chloride (NaCl) to precipitate suspended genetic material. The tubes are centrifuged at 4°C in two sequential steps, removing as much of the supernatant as possible with the resulting solids (pellet) submitted for further processing.
Bureau Veritas uses the TRIzol™ extraction method to isolate the genetic material from the pellet. Once extracted, the genetic material is stable for extended periods if frozen.
Figure 1, below, provides a summary of the Bureau Veritas method for quantifying the SARS-CoV-2 genetic material in wastewater samples.
Figure 1. Summary of the Bureau Veritas method for SARS-CoV-2 testing on wastewater samples: sample prep, extraction and quantification
PCR DETECTION AND QUANTIFICATION
RT-qPCR is a well-established approach for identifying and quantifying double stranded deoxyribonucleic acid (DNA). The SARS-CoV-2 virus contains single stranded ribonucleic acid (RNA), which requires reverse transcription to complementary DNA (cDNA). Bureau Veritas’ method combines reverse transcription, PCR amplification of the resultant cDNA and quantification in a single step. Thus, it is referred to as 1-step qPCR method. This approach offers better sensitivity and more representative results.
PCR Identification and quantification of genetic material targets genes that are specific to a particular pathogen. In this case, the Bureau Veritas method identifies and quantifies the N1 and N2 fragments of the SARS-CoV-2 N gene, which encodes the virus’s nucleocapsid protein (N1 and N2 domains).
In addition to the SARS-CoV-2 N1 and N2 fragments, each sample is also analyzed for the pepper mild mottle virus (PMMoV), as a biomarker of fecal content. More importantly, to minimize data variability inherent in wastewater samples, the mean gene copies per mL for SARS-CoV-2 result (combined N1 and N2) is normalized to the gene copies per mL of the PMMoV.
DATA REPORTING
The certificate of analysis reports presence / absence of the SARS-CoV-2 virus based on amplification of the target genes (N1 and N2) within 40 PCR cycles. Quantitative data is reported as gene copies per mL of each of the target genes as well as a mean of the gene copies per mL of the N1 and N2 results. The method’s limit of detection is 2 gene copies/mL. Amplification of virus’s genetic material beyond 40 PCR cycles is considered as “present but not quantifiable”.
DATA UTILITY
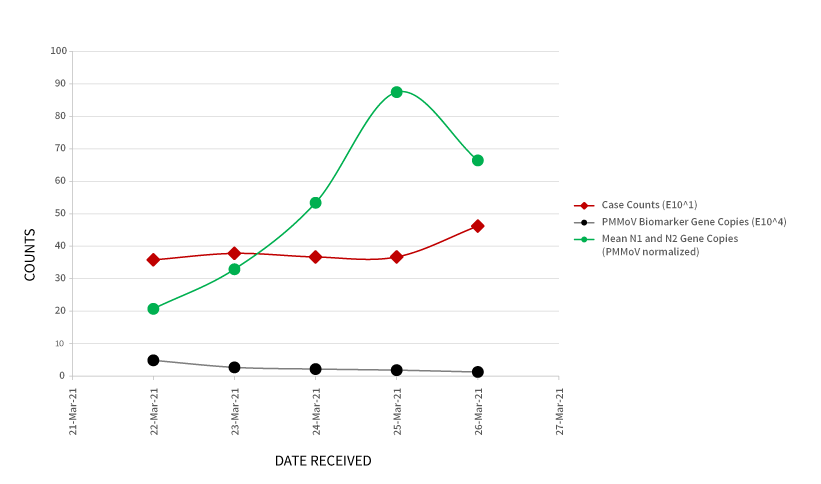
As an example of how these results can be used, Figure 2 (below) provides trending of SARS-CoV-2 wastewater data from a treatment plant in southern Ontario. In addition to the normalized gene copies per mL of the SARS-CoV- 2 virus, the graph includes associated case counts reported by the local Public Health Authority, and the gene copies per mL of the PMMoV biomarker.
Figure 2. Trending of SARS-CoV-2 genetic material (normalized data) in wastewater samples, relative to case counts and PMMoV biomarker.